Ancient DNA from the Youngest Victims of the Sacred Cenote: A Tale of Three Children and Two Babyes in the Nearby Mexican Cave and Cave
Now, ancient DNA from some of the city’s youngest victims adds to this story. A study published today in Nature analysed genomes from skulls of dozens of children and babies recovered from an underground chamber at the site in modern Mexico. They were all boys, and there were a surprising number of close relatives.
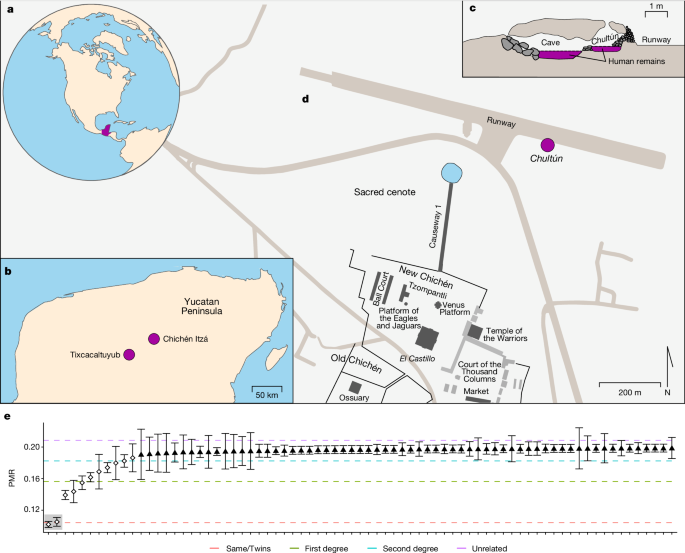
Children were found near the Sacred Cenote in an underground chamber called a chultz and an adjacent cave in the 1960s. There was no evidence of violence, but the remains were found as part of a shrine.
In the hope of identifying the sex of the remains and to glean other genetic insights, Del Castillo-Chávez teamed up with immunogeneticist Rodrigo Barquera and palaeogeneticist Johannes Krause at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, and their colleagues. The skulls of 64 individuals were obtained from the chultn.
It’s not fully clear why these children were selected for sacrifice. Isotopic analysis of their bones suggested that their plant-heavy diets — probably maize (corn) — were typical of ancient Maya. They were raised in the same way, and they had similar isotopic profiles.
It is said that it was part of preparing them for the sacrifice. “Death and sacrifice for them means something completely different to what it means to us. For them, it was a big honour to be part of this.”
Influence of cross-mapping on HLA frequencies using smartpca and OptiType for complex ancestry mapping and application to the cocoliztli epidemic
An allle that has become more common has been linked to protection against serious infections. A previous study by Krause’s team has linked the bacterium Salmonella enterica sp. Paratyphi to a sixteenth-century disease outbreak called the cocoliztli epidemic4, which killed millions of people in Mexico and beyond.
Smartpca (v.16000) from the Eigensoft package160 was used to calculate principal components (PCs) of variation in the dataset, using the options ‘lsqproject: YES’ and ‘shrinkmode: YES’. To access the continental-level ancestries of the ancient individuals were projected onto PCs using the genetic variation from 373 worldwide populations. The ancient individuals were projected onto PCs on the basis of variation from 172 Native American populations.
We applied a development version of OptiType151 on sequence data from the immune-captured libraries, mapped against a custom HLA reference panel containing 1,025 alleles with ‘common’ or ‘intermediate’ CIWD 3.0 designation (https://github.com/FRED-2/OptiType, tag GRG). The number of the best matches was set to zero. We obtained the alleles for the genes in class I and class II. Of the 882 raw allele calls, 72 (8.1%) Secondary allele predictions were overruled when it came to coverage patterns that were caused by cross-mapping. The frequencies and properties that were reported were used to assign Haplotypes. kinship and when possible. To compare the HLA frequencies between precontact Chichén Itzá and postcontact Tixcacaltuyub, we performed Fisher exact tests154 and adjusted the P values for several comparisons according to the Benjamini–Hochberg correction155 which control the false discovery rate (FDR). We use the FDR adjustment as, compared to methods such as the Bonferroni correction, the FDR approach controls for a low proportion of false positives, instead of ensuring the existence of absolutely no false positives, resulting in increased statistical power155. It has been argued74 that certain types of non-overlapping association between HLA loci may be a signature of pathogen selection. The data for every HLA locus was used for the metric 74,75,156. To provide a point of comparison for the ({f}{{\rm{adj}}}^{ }) score of each locus pair for both YCH and TIX, we generated 5,000 random permutations of the order of the total alleles at one of the loci in each pair and recalculated ({f}_{{\rm{adj}}}^{ }) for each set of randomized data. We generated distributions of possible ({f}{{\rm{adj}}}^{ }) scores for each pair of loci in the dataset that would be obtained if the alleles at those loci were associated entirely at random. The difference between the f_rmadj value and the mean of the scores calculated from the randomized study was calculated. The resulting scores allowed us to rank the HLA pairs in order of how extreme an amount of non-overlap they showed relative to entirely random associations between the same alleles (Supplementary Methods: ‘Non-overlapping associations’). To further assess the role of HLA class II alleles in resistance to S. enterica infection, we ran in silico binding predictions for the HLA-DRB1/3/4/5 alleles and -DQA1/DQB1 allele pairs found in YCH individuals and peptides derived from 18 Salmonella spp. proteins that have been previously reported to be highly immunogenic in humans (Supplementary Methods: ‘In silico binding prediction assays’) The IEDB Analysis Resource has a virtual machine image157,158. Binders were classified as strong (rank less than 2%) or weak (2% ≤ rank ≤ 10%) on the basis of the adjusted rank values recommended by the authors and previously published research82,159.
The haplogroup mapping was done to the revised Cambridge reference sequence. We used HaploGrep2 and HAPLOFIND149 to assign and confirm the corresponding haplogroups. For the Y-haplogroup assignments, we created pileups of reads for each individual which mapped to Y chromosome (Y-Chr) SNPs as listed on the ISOGG Y-DNA Haplogroup Tree (v.15.73; https://isogg.org/tree/). We then manually assigned Y-Chr haplogroups for each individual based on the most downstream SNP retrieved after evaluating the presence of upstream mutations along the Y-Chr haplogroup phylogeny (Supplementary Fig. 6). Full mtDNA haplogroups and Y-Chr genotypes can be found in Supplementary Tables 9 (YCH) and 10 (TIX).
Using an in-solution capture approach based on modified immortalized probe sequences136, target immunity genes sequences, mtDNA, Y chromosome or a panel of 1,237,207 SNPs were enriched from the total DNA in the sequencing libraries32,33,137,138,139,140. The captured library pools were split into smaller pools which were then mapped on the Hiseq platform in 75 cycles.
The libraries we built using 15 l of YCH extract were constructed to assess the authenticity of the data obtained from aDNA129. We then used 20 μl of each YCH extract to build UDG-half libraries with Illumina-specific adaptors following a modified double-stranded library preparation protocol as previously described130. The libraries we built for TIX extract were not treated with Udg. Both YCH and TIX libraries were quantified using quantitative PCR (qPCR) with the IS7 and IS8 primers in a quantification assay using a DyNAmo SYBR Green qPCR Kit (Thermo Fisher Scientific) on the LightCycler 96 platform (Roche Diagnostics).
The bone powder was decalcified and then it was taken out of the buffer at 37 C and taken to a lab where it was taken out of the buffer again. The DNA was purified from the supernatant by a silica column-based method using a silica column for high volumes assay (High Pure Viral Nucleic Acid Large Volume Kit, Roche Molecular Systems). There was 100 l of DNA in TET. One mM of tris. The library preparation128 was frozen at 20C until it was ready. In contemporary participants, 5ml of peripheral blood was collected and used to extract the DNA, using the Quick-DNA Miniprep Plus kit from Zymo Research Corporation. Our protocols are designed for short-length aDNA and to avoid biases, we sheared the DNA from modern people using an average length comparable to that of a DNA. Therefore, 50 μl of a 50 ng μl−1 dilution of each of the modern DNA samples were sheared to an average fragment length of 150 base pairs using a Covaris M220 Focused ultrasonicator (Covaris).
Direct insight into dietary trends among past populations enables the investigation of connections between diet and social status, cultural customs linked to food, environmental impacts on subsistence and perhaps even individual mobility122,123. Measurements of bone collagen δ13C and δ15N disproportionately reflect the protein component of an individual’s diet during the period of tissue formation and, to a lesser extent, the lipid and carbohydrate sources124. To reconstruct the diet of the subadult individuals from Chichén Itzá studied, we analysed bone collagen from temporal and petrous bones (Supplementary Fig. 4). A standard procedure was used to extract the human tissue. The C:N ratio is used to determine the quality of collagen preservation. The C:N ratio should range from 2.8 to 3.6 for archaeological samples, but a higher yield of more than 1% is acceptable for carbon and nitrogen values.
The portions of the petrous bone of 26 YCH samples were analysed using a standardized procedure. Collagen was extracted from the bone samples (approximately 1 g, using a modified version of the Longin method118), purified by ultrafiltration (fraction greater than 30 kD) and freeze-dried. The CO2 was burned in the analyser. The CO2 was analysed using an AMS system. The measurement of the ratio of 14C and 13C to the sample was accomplished in the AMS system. 14C-ages are normalized to δ13C = −25‰ (ref. 119) with a typical uncertainty of 2‰ and calibrated using the dataset IntCal20 and the software Oxcal (v.4.3.2)120,121.
Source: Ancient genomes reveal insights into ritual life at Chichén Itzá
Towards a Co-Responsible Model of Healthcare: Community Engagement in Tixcacaltuyub 2023
The community of Tixcacaltuyub self-identifies as a Mayan community and has been in a years-long cooperative relationship with the Chemistry and Nursing Faculties of UADY, Mérida, Yucatán, following projects investigating the relationship between health and lifestyle in the community. J.C.L.R., M.E.M., and J.C.T.R. are working together with the community to develop health interventions. As a result of these interventions, healthcare barriers and opportunities have been identified and work is now underway with the active participation of the community to implement a co-responsible model of healthcare. In April 2023, J.C.L.R. and R.B. visited Tixcacaltuyub to hold meetings with participants and elementary, high school and UADY students to return the results of the genetic findings and to collect community feedback, including how they reconcile these results with their own views. feedback from these engagements was included in the final manuscript. There is a non-Peerreviewed, Spanish translation of the main manuscript in our strategies for outreach.