Genome-wide variant effect prediction using deep learning-derived splice scores: Saturation genome editing-based clinical classification of BRCA2 variants
Rentzsch, P., Schubach, M., Shendure, J. & Kircher, M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. In the Genome Med. 31 will be in the years 2021.
Tavtigian, S. V., Deffenbaugh, A. M., Yin, L., Judkins, T., Scholl, T., Samollow, P. B., de Silva, D., Zharkikh, A. & Thomas, A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. It was Genet. A total of 43, 295, and 301 were recorded in 2005.
Source: Saturation genome editing-based clinical classification of BRCA2 variants
bbac074: an automated pipeline for in silico saturation mutagenesis of protein structures and ensembles. AACR Project GENIE Biopharma Collaboration. cBioPortal. Cancer Res. 83,
Tiberti, M. MutateX: an automated pipeline for in silico saturation mutagenesis of protein structures and structural ensembles. Brief. The bbac074 was the 23rd book in the bbac series.
de Bruijn, I. et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 83 was published in 203.
Sorrentino, et al. The integration of VarSome into a bioinformatic platform for automated ACMG interpretation is in the works. Rev. Med. The use of the word pharma. There is a report in the edition of the siac 25, 1–6 in2021.
Walker, L. C. et al. The ACMG/AMP framework can be used to collect evidence related to predicted and observed impact on splicing. Am. It is J. Hum. It’s called genes. 1086, 1088, 1089, 1098, 1099, 1099, 1099, 1099.
Easton, D. F. et al. There are 1,433 sequence variant of unknown clinical significance in the breast cancer-predisposition genes. Am. That’s a hum. Genet. 81, 873–832.
Finding disease-causal variant in a wealth ofgenomic data with needles in stacks. Nat. Rev. Genet. 12, 628–640 (2011).
Tabet, D., Parikh, V., Mali, P., Roth, F. P. & Claussnitzer, M. Scalable functional assays for the interpretation of human genetic variation. Rev. Genet. 5, 19.15–19.25 (22)
Findlay, G. M., Boyle, E. A., Hause, R. J., Klein, J. C. & Shendure, J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123 (2014).
Functional Evaluation of a Sample of BRCA2 and DSS1 Variants in the Sri Lankan Genome Using VarCall Models
N., Sirisena, and other people. A mouse-based test is used for functional evaluation of the five unclassified variants found in the Sri Lankan cohort. The Breast Cancer Res. 22 and 43 were published in 2020.
A review of cancer genes using a base editing screen with efficiency correction. The genomes were included in the 81 in the Genome Biol.
Mishra, A. P. et al. BRCA2–DSS1 interaction is dispensable for RAD51 recruitment at replication-induced and meiotic DNA double strand breaks. Nat. Commun. 13, 1751 went into effect in 1992.
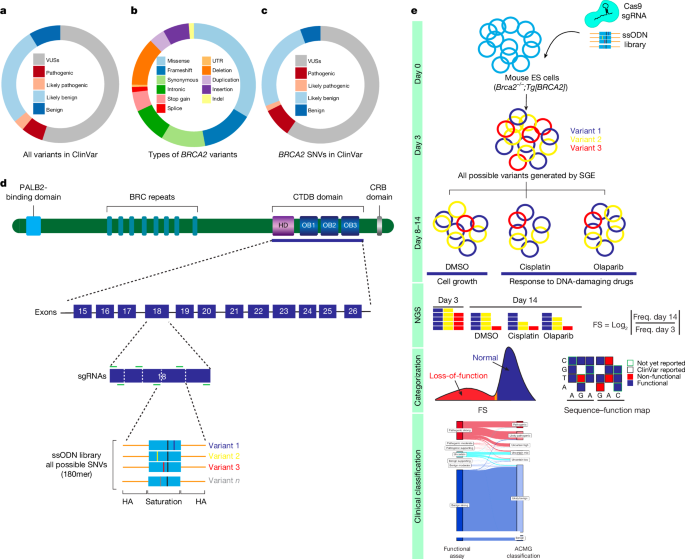
The replica-level variant frequencies were calculated at the three time points by summing the variant read count by the replicate total for each exon. The positionality effect was estimated using the ratio between D0 and D5 read counts. The log ratio of the D14 and D0 was adjusted using the VarCall model37. VarCall is a class of Bayesian hierarchical model with context-specific measurement models that embed a Gaussian two-component mixture model for the variant effects. The previous analysis of BRCA2 variant8 was used in the formula that was used here. Variants were each assigned a binary indicator of pathogenicity status: deterministically if assumed known and probabilistically if not. The silent and nonsense variants were assumed to be benign. The model was adjusted to fit for time by including replicate by exon level location, random effects and t-distributed error terms. MCMC was used to calculate the fit of the VarCall model. The computations were done in the R programming language. AlphaMissense predicted a frequency of 0.23 for variants in the DNA-binding region which was used to derive a prior probability of 0.2 for them. The Bayes factor was used to determine the pathogenicity of each variant. The Bayes factor is based on the strength of evidence of benign or pathogenic level and is determined by the ACMG–AMP guidelines. The full analysis can be found in the Supplementary Methods.
Richards, S. et al. The guidelines for interpretation of sequence variant were created by the American College of Medical Genetics and Genomics and the Association forMolecular Pathology. The genes are Genet. Med. 17 pp. 423–423.
Biswas, K. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood 118, 2435–2724.
Arnaudi, M. et al. MAVISp: multi-layered assessment of variants by structure for proteins. Preprint at bioRxiv https://doi.org/10.1101/2022.10.22.513328 (2023).
FASTQ files of sequenced samples from Illumina MiSeq or NextSeq assays were trimmed for adapter sequences using cutadapt (v.3.5). Single reads were converted into the Pairs-end reads by SeqPrep. The human reference genomes were aligned with the single reads using bwa-mem. Following alignment, the custom-developed tool CountReads was used for DNA-sequencing data analyses, with a particular focus on the identification and characterization of mutations. CountReads included the preparation of a lot of different kinds of reads, from a reference to a read. The method also differentiated between variant types and confirmed the presence of specific variants and aggregated and reported variant data. A variant call format file was annotated using CAVA35. The SpliceAI tool (v.1.3.1)36 was utilized to evaluate splicing effects associated with all observed SNVs.
The log2 ratio between the frequency of D14 and D0 read counts was used to measure the depletion or enrichment effect for each variant. The comparison between experimental D0 and D5 was used for positional adjustment using a Loess transformation6. Variants with under-represented read counts (<10) at D0 and D5 were excluded from further analysis. log2 ratios of variants were linearly scaled within each exon across replicate experiments relative to median silent and median nonsense SNV values. The average score for each variant was calculated using all the non-missing values. Linear scaling was used to amplify scores in exons using median synonymous and nonsense values. After completion of all data cleaning and quality control, a raw functional score was available for 6,959 SNVs (Supplementary Table 3).
The missense alterations were mapped in the database using the PyMol software. The identifier 1MJE is a source file of the Protein Data Bank. Three-dimensional structural modelling was based on the crystal structure of a BRCA2–DSS1–ssDNA complex39.
BRCA2 amino-acid sequences were obtained from Align-GVGD (http://agvgd.hci.utah.edu/). Ten species were used for the alignments, including Homo sapiens, canis familiaris, Macaca mulatta, Rattus norvegicus, and Canis familiaris. The analyses were done on the affected amino-acids. Align-GVGD26, AlphaMissense27 and Bayes-Del40 were used for in silico pathogenicity prediction.
SGE functional results were compared with those from other studies, including a BRCA2-deficient cell-based HDR assay7, a BRCA2-deficient cell line–based drug assay24, a prime-editing-based SGE study16 and a mouse embryonic-stem-cell-based functional analysis25.
The ACMG–AMP rule-based framework combines evidence from population, computational and predictive, segregation, functional, and other data, with each contributing source weighted as very strong (PVS1), strong (PS1, PS2, PS3 and PS4), moderate (PM1, PM2, PM3, PM4, PM5 and PM6) or supporting (PP1, PP2, PP3, PP4 and PP5) evidence for pathogenic effects, or stand-alone (BA1), strong (BS1, BS2, BS3 and BS4) or supporting (BP1, BP2, BP3, BP4, BP5, BP6 and BP7) for benign effects. LB, pathogenic, LP and VUS9 are variant classifications from the combined data. In this study, ACMG–AMP scoring rules established by the ClinGen BRCA1/2 VCEP were used for clinical classification of BRCA2 DBD SNVs. The PS3/BS 3 rule allowed the integration of the functional data into the Clin Gen–ACMG–AMP BRCA1/2 VCEP classification model. The values for functional evidence were capped at +4 and -4 on the log scale to avoid classification with functional evidence alone. The study was approved by the Western Institutional Review Board, which exempted review of the clinical testing cohort, and by the Mayo Clinic Institutional Review Board (21-008216). Detailed ACMG–AMP criteria used in this study are provided in the Supplementary Methods.
The LOH status for breast, ovarian, and pancreatic tumours carrying germline BRCA2DBD variant was acquired from the sample from the IMPACT dataset32. The FACETS was employed to determine LOH from matched tumour–normal pairs. Only tumour samples with >40% tumour content were included in the analysis.
Integrated DNA Technologies Evaluation and Amplification of HAP1 Cells (Horizon Discovery) in IMDM for RNA Synthesis (SGE Experiments)
The consent of the study participants is given to the data that is shown in the paper after the approval from the institutional review boards.
HAP1 cells (Horizon Discovery) were maintained in IMDM with 10% FBS and 1% penicillin–streptomycin. For haploidy sorting, 1 × 10−7 The HAP1 cells were resuspended and sorted at 4 C. HAP1 cells were transfected using Turbofectin 8.0 (Origene). Integrated DNA Technologies produced all the primers and oligonucleotides.
Multiple sgRNAs with predicted high editing efficiency in HAP1 cells were evaluated in SGE experiments, and optimal sgRNAs were selected (Supplementary Table 1). In each SGE experiment, 5 million haploid-sorted HAP1 cells were co-transfected with 4 mg of the target-specific variant library and 16 mg of the Cas9–sgRNA targeting construct. Cells were found in puromycin for 3 days. Cells were collected at D0, D5 and D14 after being transfectioned, and gDNA was retrieved using a Monarch Genomic DNA Purification kit. Target regions were amplified by the method of genetic modification. All PCR reactions were performed in 50 μl reactions using Q5 High-Fidelity 2× master mix (New England Biolabs, M0492L). Supplementary Table 2 includes primer information for amplification. All reactions were cleaned and concentrated using Ampure XP beads before sequencing for 150 cycles on an Illumina MiSeq (approximately 5 million reads per run) or NextSeq (approximately 30 million reads per run) instrument. The base calls were made using the instrument control software.