The Father’s Microbiome as a Modified Fermion-Induced Effect on Placenta Development in Mammalian Embryos
In mammalian species that form placentas, embryonic development and subsequent fetal growth depend on the genetic contributions and related signals carried in the egg and the sperm, with roles for the placenta, the maternal host tissues and the external environment. The influence of gases, vitamins and hormones is caused by the exchange of gases. They are also affected by exposure to microbial or viral disease-causing agents and to inflammation, toxic compounds and social and behavioural stressors. The integration of these signals determines the outcome of pregnancy, and adverse influences can compromise fetal and maternal health and lifespan.
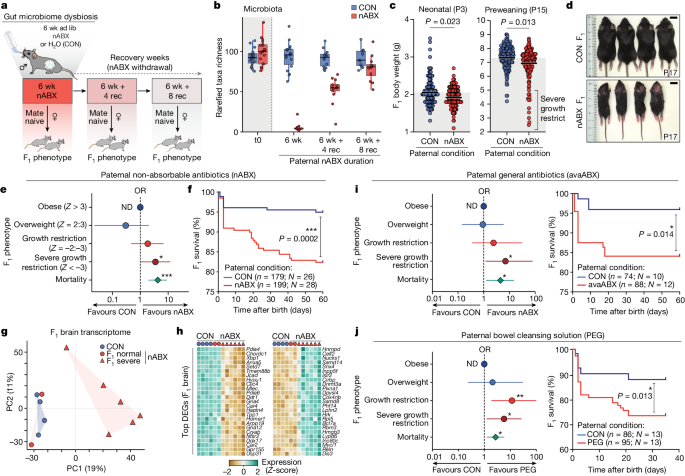
These exciting observations establish a link between the paternal gut microbiota, sperm RNA content and pregnancy outcome. Although the mechanisms linking altered sperm biology with changes in the offspring and placenta and with altered gene expression remain to be unravelled, this line of investigation highlights antibiotic-mediated disruption of the paternal gut microbiota as a previously unknown mode of a sperm-mediated effect on fetal development and offspring health. The work suggests that the Father’s Microbiome may contribute to the health of his unborn child in a way never before seen in humans.
Using a Fisher’s exact test, a class and enrichment analysis was calculated for the treated and control groups. All P values were FDR-corrected for several hypotheses testing using the Benjamini–Hochberg procedure (stats::p.adjust function in R with BH parameter).
Statistical significance of the feature intensity differences was assessed using a two-sided t-test (stats::t.test function in R) of log-scaled data and P values were FDR-corrected for several hypotheses testing using the Benjamini–Hochberg procedure (stats::p.adjust function in R with BH parameter). A visualized P value was displayed using a Volcano plot. Mass, retention time, HMDB annotation, class, superclass and composite spectrum were retrieved for all features showing significantly different intensity between the treated and the control group and an absolute foldchange greater than 2.
In order to get theAnnotation, you have to search for the exact same mass or 20ppm window from a mono-isotopic mass of a metabolite. Several annotations were retrieved when present. Only features being annotated were retained for the downstream analysis. The superclass and metabolite class were retrieved for each feature.
Cutadapt was used to remove the first ten bases of both reads, followed by trimming the alignment reads. There are Variants that were selected to remove the individuals with the QUAL of less than 30 and an allele Frequency of less than 0.2 in more than one individual. An updated version of the functional region was annotated. Structural variations were called with the Delly2 call.
Raw fastq sequences were quality- and adaptor-trimmed using Trim Galore (0.4.3.1) and reads aligned to mm10 using Bismark (0.20.0), discarding the first 8 bp from the 5′ end and the last 2 bp from the 3′ of a single-end reads. The katasine status was retrieved from the mapped reads with the help of the Bimark tool. Seqmonk software used five biological independent replicate datasets for each condition to analyse thegenome-wide methylation calls. The first thing the genome was binned into was a set of sliding tiles containing 50 consecutive CpGs. The minimum of 10 reads was used to run a read-depth sensitive Logistic Regression (P(adj) 0.05), which identified DMRs. The methylation level at specific genomic features (for example, imprints) was calculated using the DNA methylation pipeline in Seqmonk over target features.
The negative binomial test was used to find significant differences after Benjamani–Hochberg multiple test correction.
nABX, CON and IVF using seqmonk for gene-length-adjusted transcriptome analysis and genome-strategy analysis
Quality was trimmed using Trim Galore (4.3.1, -phred33, quality 20, stringency 1,e 0.1,length 20). The reads with a MAPQ score less than 20 were discarded to ensure only unique-mapped high-quality. The data were quantified using the RNA-seq quantification pipeline for directional libraries in seqmonk software to generate log2 reads per million (RPM) or gene-length-adjusted (RPKM) gene expression values.
Principle component analyses of transcriptomes were computed in seqmonk and R statistical software using all expressed genes as input. These were defined as having an RPKM > 0.1 in at least two replicates across all assayed samples.
The DEGs were determined using the DESeq2 package, inputting raw mapping counts and applying a multiple-testing adjusted P value. An extra fold change was applied to generate DEGs.
For each cell type identified in the dataset, the FindMarkers function was used to identify DEGs between nABX and CON cells using logfc.threshold = 0.25 and p_val_adj < 0.01.
The annotated dataset used uniquely expressed marker genes. As above, clusters showing no uniquely expressed genes were ignored. The germ cells were separated into different Seurat objects and clustered separately, to define more fine-grained annotations.
The nABX and CON samples were then integrated using the canonical correlation analysis approach at default parameter settings as recommended by the Seurat package.
Testes-to-body weight ratio: to assess the effect of nABX treatment, the body weights of sires treated for 6 weeks were measured and compared with those of age-matched CON sires. The impact on reproductive organs were assessed by weighing testes from CON and nABX- treated mice and calculating the testes-to-body weight ratio difference between the two groups.
Natural and artificial (IVF) methods were used to generate F1 offspring. The coding system was used to make sure that researchers were double- blind to the paternal treatment regime.
To separate the potential indirect impacts of keeping females with a male with a yeasty smell from keeping females with a male with no yeasty smell, we used a co strategy. The virgin female was put in with the male with either 6 weeks of previous n ABX exposure or a control male during the day, recapitulating parental co-exposure. Males were removed from the cages at 16:00 to prevent mating during the night, with females remaining in the male cages to maximize the potential to detect microbial transfer or coprophagic effects. All females with vaginal plugs were excluded from the study, while all females with vaginal plugs were held in the male cages. In the 4 day cohousing period, mice were maintained in standard optimized environmental conditions with ad libitum access to food and water. All plug-negative females that had been exposed to nABX and CON males were matched with the 11-week old C57BL/6J male mice to find out if there was any indirect effects on previous nABX male exposure.
The peripheral part of the pre-incubation drops contains the most motile sperm, and that is where the 10 l of sperm suspension was taken. Aliquoted sperm were then transferred either from CON mice or nABX-treated mice into the centre of the drops in the fertilization dish containing oocytes. After insemination of the oocytes with sperm, sperm–oocyte fertilization dishes were cultured at 37 °C under 5% CO2 for 3–4 h. After 3–4 h of incubation, the presumptive zygotes were washed three to four times with 150 μl drops of M2 medium and cultured overnight in four-well NUNC plates with 500 μl of KSOM medium supplemented with amino acids (KSOMaa) at 37 °C under 5% CO2 and 5% O2 incubator. The fertilized two-cell stage embryos were taken to the oviducts of pseudopregnant CD1 and C57BL/6J female surrogate mice.
Fresh sperm was prepared from cauda epididymis from 11-week old mice which had their adipose tissue and blood cleaned away. Subsequently, cauda epididymis was transferred into a centre-well organ culture dish (Falcon) containing 120 μl of cryoprotective agent (CPA). In order to release the sperm, 5 to 6 incisions were made across each cauda epididymis with scissors. Fresh sperm were pre-incubated in CPA for 3 min at 37 °C with gentle stirring of the Petri dish every minute. A 10 μl sperm suspension was transferred into the centre drop of 90 μl of TYH-MBCD medium and the fresh sperm were capacitated by incubation for at least 10–30 min at 37 °C in the 5% CO2 incubator. Dishes remained undisturbed until the sperm were moving rapidly and equilibrated in the medium.
Seminal vesicles were collected in a laminar flow-hood as described previously57 with slight modifications. To prevent crosscontamination, one experimental animal was placed in the hood. To ensure maximum sterilization, the experimenter wore a sterile gown, gloves, mask and scrubbed their hands with 70% ethanol and followed the following procedures. In brief, the mice were placed in a laminar flow-hood that had been previously cleaned with 70% ethanol and 2% Virkon followed by 2 h of ultraviolet radiation. mice were euthanized, and their abdominal areas were cleaned with 70% solution and incised with scissors after being euthanized. Seminal vesicles were then excised using new sterilized tweezers and scissors and placed in a sterilized 2 ml Eppendorf tube, snap-frozen in dry ice and stored at −80 °C until DNA extraction for seminal fluid microbiome profiling. The sample collection only had dissection tools used for once per mouse.
The blood samples were taken from the microvettes, which were placed at 5000rpm for 10 minutes and then cooled down to -40 degrees.
The placenta samples were fixed in 4% paraformaldehyde at a temperature of 4 C after being embedded in paraffin. Sectioned samples were mounted on slides and dried on a hot plate. The dried sections were put to bed overnight at 4 C and then exposed to heat-inducible antigen retrieval using a citrate buffer. The catalogue no.14-1441-81 was detected by anti-Rat-Alexa Fluor 568. A-11077 has 4 gml. There was a counterstained of 4′s6, diamidino-2-phenylindole, according to the catalogue no. D1306; 5 μg ml−1). Slides were washed before being mounted with ProLong Glass (Thermo Fisher Scientific; catalogue no. P36983) and imaged on a Leica THUNDER Imager Live Cell with a ×20 objective (numerical aperture = 0.8). Qupath was used to measure the areas of labyrinth zone.
ELISA kits were used to determine the concentration of a specific target protein in biological samples. Frozen plasma and testis samples were used for the measurement of leptin (Lep) in F0 sire males, whereas F1 placenta tissues were used for the measurement of placental growth factor (Plgf) and soluble fms-like tyrosine kinase-1 (sFlt1). Mouse Leptin ELISA kit MOB00, mouse PlGF-2 The person is called ELISA. Following the instruction from the manufacturer, the kit mp200 and mouse VEGFR1/Flt-1 kit MVR 100 was used for the quantification of sFlt1. All samples were prepared as per the instructions given on the kits and all standards and control were duplicated in order to perform more accurate analyses. The model curve fit gave the data to quantify the target concentrations.
Source: Paternal microbiome perturbations impact offspring fitness
Metabolomics: Screening for antibiotic residues and sample preparation for the quantification of antibiotics in testis tissues and the circulatory system
We had many different approaches used to screen for antibiotic residues and also to test if nABX is present in the tissues or the circulatory system.
Samples for the quantification of antibiotics in testis tissues were prepared as described in the section on Metabolomics below. A 20 l sample was resuspended in water and a 10 l sample was injected into a mass spectrometer. The following ranges of injected amounts were used as the basis for the curves: bacteriocin ( 8.6–0.0 pmol) and neomycin (7.5–112.5 pmol). Quantification of antibiotics was performed using the MassHunter Quantitative Analysis Software (Agilent Technologies, v.10.0) and limit of detection was determined by linear regression using R.
PremiTest kit (R-Biopharm) was used for the determination of nABX or avaABX residues in testis samples stored at −80 °C, which was performed in accordance with the manufacturer’s instructions for use in meat. The test is based on a bacterium that can be sensitive to antibiotics and other compounds. After the testis were frozen, they were washed in PBS and then homogenized with 200 l of tissue grinder. In this case, nABX cocktail was used as a positive control at the minimum detection limit while sterilified water was used as a negative control. 100 l was pipetted onto the agar in the Ampoule and given a chance to stand at room temperature for 20 minutes prior to prediffusion. The sample solution was washed twice with water and then covered with foil to make it less prone to evaporation after prediffusion. Subsequently, the test ampules were incubated in Eppendorf block heater at 64 ± 0.5 °C for approximately 3–3.5 h until the negative control turned from purple to yellow. The kit bicolour indicator was used to interpret the results after the block heatersampoules were removed.
The testis-to-body weight ratio and standard male fertility parameters are characterized on the basis of a quantitative descriptive analysis.
The samples were kept at 80 C until their 16S rRNAs were processed. Microbial DNA from a faecal specimen could be found in the QIamp PowerFecal Pro DNA Kit (QIAGEN) according to the instructions. In order to quickly mix faecal samples processed with the kit, a PowerBead beat tube was added and the samples were rapidly homogenized on the mixer. When cells are lysed and potential inhibitors removed, total genomic DNA is washed and eluted before being used in gut microbiota profiling.
The relative abundance of 16S rRNA genes was determined by using a standard curve and serial dilutions of the control sample.
Targeted amplification of the 16S rRNA V4 region (primer sequences F515 5′-GTGCCAGCMGCCGCGGTAA-3′ and R806 5′-GGACTACHVGGGTWTCTAAT-3′ (ref. minor modifications from the manufacturer’s instructions were made to the protocol performed using the KAPA HiFi HotStartPCR mix (Roche). PCR products were pooled, purified using size-selective SPRIselect magnetic beads (0.8 left-sized) and then sequenced at 2 × 250 base pairs (bp) on an Illumina MiSeq (Illumina) at the Genomics Core Facility, European Molecular Biology Laboratory, Heidelberg.
The reads were trimmed, denoised and then filters to remove the chimaeric PCR artefacts. 61). The reads were mapped to a pre-clustered reference set of 16S rRNAs at 98% similarity using the open-reference approach and the amplicon sequence variations were then clustered into operational taxonomic units. Reads that did not confidently map were aligned to bacterial and archaeal secondary structure-aware rRNA models using Infernal63 and clustered into OTUs with 98% average linkage using hpc-clust64, as described previously65. The OtU count tables were noise filters that claimed that samples with at least 1000 reads and taxa were prevalent in at least two samples, but they still removed almost all of the spurious OTUs from the dataset.
The average values of 100 rarefaction iterations to 5000 reads per sample were calculated by combining Otu richness, exponential Shannon entropy and inverse Simpson index. Between-sample community diversity was calculated as Bray–Curtis dissimilarity. Trends in community composition were quantified by means of ordination methods,distance-based analysis and permutational multivariate analysis of variance, implementing the R package vegan68.
The Smart-Seq protocol was used to carry out the single-embryo transcript analysis. 70. For quality control and data preprocessing, raw reads were aligned using STAR v.2.7.10a (ref. 71) at default parameter settings to mm10 (GRCm38) primary genome assembly. The data was measured using the featureCounts module of Subread. The expression and PCA were performed in R. Differential expression was performed by passing the raw counts into the DESeq2 (v.1.34.0) package73. The vst function was used to normalize the counts at default settings and the top 500 variable genes were used for PCA. The Metascape web application was used to infer the enrichment of genes. The list of genes was uploaded to Metascape as a text file. The option was selected after setting the parameters of the analysis to M. musculus.
Testicular single-cells resuspended in PBS + 0.04% BSA were adjusted at a concentration of about 1,100 cells μl−1 for loading on the 10x Chromium chip. As per the instructions, cell capturing and library preparation was carried out. v.3.1 (Dual Index) User Guide). After 12 cycles of library amplification, about 6,000 cells were targeted for capture. The resultant libraries were size selected, pooled and sequenced using Illumina NextSeq 2000 P2 flowcell (100CYC) is a protocol.
The purification of sperm genomic DNA is accomplished with the help of the DNeasy Blood & Tissue Kit. The DNA extraction was performed as follows. 100 l of DNA was being taken out of frozen pure sperm samples at 80 C. Tris Cl pH 8, 20 mM 200 mM is EDTA. After the sample was prepared, it was put into the Eppendorf thermomixer at 56 C and subjected to complete sperm lysis for 1 h. The purification of total DNA from animal sperm is accomplished using the user-developed protocol DY03 (QIAGEN). BS-Seq libraries were constructed according to the manufacturer’s instructions using TruSeq DNA Methylation Library Prep Kit (Illumina). Amplified libraries were multiplexed and sequenced on an Illumina NextSeq 500 (PE75).
The gDNA was taken from the normal or SGR offspring of F1 using the manufacturer’s instructions. The quantity and quality of DNA were compared to make sure they were within the integrity limit. For library preparation NEBNext Ultra II DNA Library Prep kit was used, following all manufacturer’s guidelines. Illumina Hisq4000 (PE150) was used to provide the appropriate depth and resolution for the libraries.
Source: Paternal microbiome perturbations impact offspring fitness
Mass calibration and alignment of LC-MS data in Fisher Scientific using MassHunter Qualitative Analysis software (Agilent, v.15.1)
Fisher Scientific purchased all the chemicals for the LC–MS analysis. The online mass calibration standards were purchased from a company.
The acquired LC–MS data was analyzed using the MassHunter Qualitative Analysis Software. The following settings were applied: peak filter of absolute height: 5,000 counts, limit assigned charge states to 1, only ±H+ charged molecules were included with compound quality scores greater than 80%. Peak alignment and identification were carried out using Mass Profiler Professional (Agilent, v.15.1) with default parameters: mass tolerance of 2 mDa or 20 ppm and retention time tolerance of 0.2 min or 2%. The features were aligned and exported as a.csv file.
Source: Paternal microbiome perturbations impact offspring fitness
Nested t-tests for intergenerational studies in the presence of individual offspring, and a statistical analysis with Seurat 6.1.2
The data wasanalyzed with graphical software and R.3.6.2. The number of uniquely exposed fathers is the basis of the replica number, which is influenced by the number of offspring which is greater. Thus, although we typically generate n > 150 offspring per experiment to robustly capture effect size and partially penetrant phenotypes, statistically speaking each individual derived from the same father is hierarchically nested into a single N value. For example a nested t-test compares the means of two groups, for which there was a nested factor in the treatment groups (shared father amongst each litter). This is necessary as using individual offspring as independent variables in intergenerational studies will lead to inflated alpha error rates and spurious significance. Two-tailed unpaired t-test was used to analysed the testes to body weight ratio, fetoplacental ratio and labyrinth zone. The odds ratios and the CIs were calculated using the Baptista–Pike method and the chi-squared test. The Kaplan–Meier method was used to derive survival analysis curves. There was an accepted curve fit of R2 > 0.99 in the Hyperbola model.
10x Genomics Cellranger 6.1.2 contains the count module that aligned raw reads to the transcriptome assembly with default parameters.
The Seurat package 76 was used to perform all subsequent steps. The number of unique genes, the number of unique Ribosomal cells and the number of unique mitochondria were all considered before cells were sieved. The sample were clustered using the default Seurat clustering approach. The clusters that had no uniquely expressed genes were discarded.
Source: Paternal microbiome perturbations impact offspring fitness
Intergenerational effects of maternal testicular leptin deficiency in a C57BL/6J ob/ob mouse model
All experiments involving mice were carried out in accordance with the approved protocol and guidelines by the laboratory animal management and ethics committee of the European Molecular Biology Laboratory (EMBL) under license no. The Italian Ministry of Health is authorized under this code. 308/2021-PR. The inbredC57BL/6J strain was the primary mouse model while the CD1 IGS or C57BL/6J dams were used as surrogate mothers. C57BL/6J mice deficient in leptin (ob/ob mice) were used to study the intergenerational effects of paternal testicular leptin deficiency. All mice were housed under a light/dark cycle, which allowed them to get their regular diet and water. Standard chow contained 18.5% protein, 5.3% fat, 4% fibre and other nutritional additives in pellet form and is suitable for long-term maintenance, breeding, lactation and gestation periods (NFM18, Mucedola).