The Genetics of Greenland’s Greenlandic ancestors: A systematics study of the environmental and ecological effects on DNA sequences
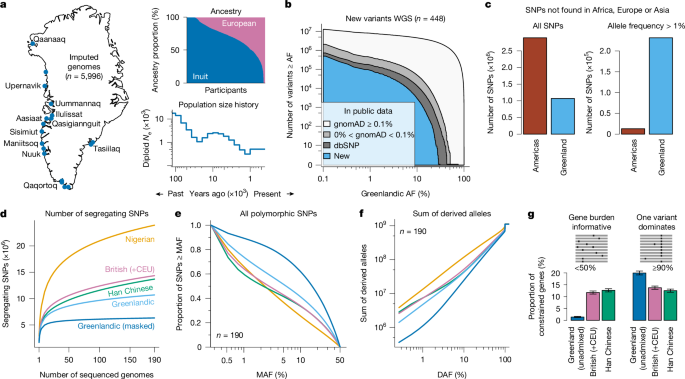
An analysis of the genomes of nearly 6,000 Greenlandic people suggests that their Inuit ancestors rarely moved around after settling the island around 1,000 years ago. People from parts of Greenland are more likely to have genetic diseases than people in other parts of the world.
The study was approved by the Scientific Ethics Committee in Greenland. No. 2011–056978), project 2014-08 (ref. no. 2014-098017), project 2017-5582, project 2015–22 (ref. The project was conducted in accordance with the Declaration of Helsinki, second revision. All participants gave written informed consent.
Polygenicity in Greenland: assessment of the inter99 data using the Infinium Express omni 24 v.1.3 chip with phased WGS and IMPUTE
6,182 people from the Inter99 cohort were included in the data for the assessment of polygenic scores, which were done using the Infinium Express omni 24 v.1.3 chip. Genotypes were called using GenCall in GenomeStudio (v.2011.1; Illumina) and quality control was performed according to a standard procedure as previously described59. Data were imputed using the Haplotype Reference Consortium reference panel on the Michigan imputation server.
To prepare a good reference panel for the downstream imputation analyses, we phased the WGS data using Shapeit2 (ref. 51) with trio and duo information. The sites that were found to be polymorphic are included in the panel. Moreover, we prepared another reference panel consisting of all participants in the 1KG Project, which we called the 1KG panel. For this panel we included all overlapping sites with the Greenlandic panel and all sites with a MAF > 1% in the populations with East Asian and European ancestry (CDX, CEU, CHB, CHS, GBR, TSI and IBS52). We then imputed the MEGA-chip data with IMPUTE2 (refs. You can use the option ‘-merge_ref_panels’ to improve imputation performance. We combined the imputed and phased data with the phased WGS data into two datasets, which resulted in 5,996 participants. Quality control on the variants, as noted in each analysis, was done for downstream analyses. IMPUTE v.2 was used to investigate the improvement of imputation accuracy by comparing it with the 1KG reference panel and showing only imputation performance on the overlap sites.
Source: Genetic architecture in Greenland is shaped by demography, structure and selection
A Neural Network Framework for Fine-Scale Analysis of Inuit Ancestry Using OlinkAnalyze R and Haplotype Clustering
Body mass index and waist- hip ratio were calculated after measuring the height, weight, systolic blood pressure, and hip and waist circumference. All IHIT participants above 18 years, B99 participants above 35 years and a subset of B2018 participants underwent an oral glucose tolerance test, where blood samples were drawn after an overnight fast of at least 8 h, and at 30 min (only for B2018) and 2 h after receiving 75 g glucose. Plasma glucose was measured at fasting, 30 min and 2 h, and haemoglobin 1Ac at fasting, as previously described56. LDL-cholesterol was calculated and the concentrations of cholesterol and high-density lipoprotein cholesterol were measured. Type 2 diabetes was defined based on the World Health Organization 1999 criteria57 and controls were defined as normal glucose tolerant based on the oral glucose tolerance test data.
The data for the participants used for the quantitative trait profiling is from the ref. 31. Using the Olink Target 96 Inflammation and Cardiovascular II panels, relative plasma levels of 184 proteins were measured in 3,732 participants across the population surveys. The 2 batches were bridged and normalized based on 16 control samples using the OlinkAnalyze R package (https://cran.r-project.org/web/packages/OlinkAnalyze/index.html). Normalized protein expression values on a log2 scale were inverse-rank normalized, including normalized protein expression data below the limit of detection. The samples were not included with the quality control warning.
Inuit and European admixture proportions were calculated using the software ADMIXTURE60 There are a subset of variants with 5% and less that 2% missingness, as well as a subset with R2 > 0.8. 61).
The neural network framework HaploNet33 was used for fine structure analysis of the Inuit ancestry. First we used window-based haplotype clustering. A window size of 1,024 SNPs was used to generate haplotype cluster likelihoods for all samples, which we leveraged to infer fine-scale population structure through both ancestry estimation and principal component analysis. We used multiple seeds to ensure that the expectation maximization method of HaploNet had made it to the final stage. The convergence criterion was defined as having two runs within five log-likelihood units of the best seed. Inuit and European ancestry were assumed to be reflected in the two ancestry sources.
Next, we used HaploNet Fatash to infer the local ancestry of the haplotypes. Fatash obtains posterior probabilities of the local ancestry per genomic window per haplotype based on haplotype likelihoods and genome-wide admixture proportion estimates obtained from HaploNet train and HaploNet admix. A hidden model with instantaneous rate change is the basis of the model. Fatak is a submodule in the HaploNet software suite. Local ancestry tracts were inferred using three different window sizes (1,024; 512 and 256 variants) to increase accuracy proximate to recombination events. If the fit was more than 50 log-likelihood units we used the smaller windows.
Reference and alternative allele counts were counted using Plink v.1.9.0 (ref. The projected number of participants using the formula binom(m,j) is 61. × binom(n − m, k − j)/binom(n,k), where k is the observed number of alternative alleles, n is the number of total alleles, m is the number of alleles to project to (that is, two times the number of participants) and j is the site frequency spectra (SFS)-bin. Each site gets a probability that there would be alternative all genes observed in a sub sample. The probabilities were summed across variants and folded to get the folded SFS.
To measure the number of segregating SNPs as a function of the number of participants sequenced, we projected the SFS to the wanted number of participants, folded the SFS and summed across all the non-zero SFS-bins. We can get segregating SNPs for all sub samples of participants from our data.
For continuous traits under the additive model, the variance explained was calculated as PVE = β2add 2AF(1 − AF), where βadd is the additive effect size and AF is the allele frequency of the effect allele69. For the model variance that is explained is calculated as per the formula shown below with rec and Fhom being the effect size and Fhom being the expected amount of homozygges. This method was used for plotting as we could calculate variance explained CIs using the same formulas but with the lower and upper CI for the effect size. For choosing the variants with more than 1% variance explained we used the formula PVE = β2/(β2 + SE(β)2 × N), where SE(β) is the standard error of β and N is the number of participants70. The liability-scale variance was calculated using the R package Mangrove.
We estimated relatedness by using a set of genetic variant that were identified through a filter, along with the inferred proportions as input. For each pair, NGSremix calculates pairwise relatedness as the fraction of loci sharing zero, one or two alleles identical by descent (represented by k0, k1 and k2, respectively). Parent–offspring pairs were defined as relationships with k1 + k2 > 0.85 and k1> 0.75 and the parent was inferred from the age of the participants. There were full sibling pairs with k1 0.7 and k2 0.1. Out of 5,828 people with age and location, we identified 1,727 parent–offspring and 1,841 full sibling relationships. Relationships were normalized to the number of possible pairs. The number of possible pairs in the relationship is calculated as the number of participants in region 1 and 2. The number of potential parent–offspring relationships was calculated in the region. nregion1 1 Within region, the number of possible pairs of full sibling relationships was calculated as npossible(1,1) 1 equals (nregion1 1)/2.
To calculate the expected frequency of homozygous carriers with the current fine structure, fhom(structure), we estimated the regional allele frequencies, AFregion, based on sample location, calculated the expected number of homozygous carriers in each region as nhom(region) = nregion × AF2region, calculated the total sum of homozygous carriers, nhom = nhom(region1) + nhom(region2) + ··· + nhom(region8) and divided with the total number of participants fhom(structure) = nhom/ntotal. The expected frequencies of different types of carriers were estimated using fhom(panmictic) as the base. CIs of the homozygous frequencies were estimated as the s.d. of the frequency estimates from 10,000 bootstrap samples.
To estimate variant age, we made 10,000 branch length samples of the local tree from the estimated ARG using the provided SampleBranchLengths script from Relate. Note that Relate does not allow for changes in the tree topology when resampling. The time was taken to the most recent common ancestor for both the variant as well as the preceding one. This yields a minimum and maximum age of the variant measured in generations for each branch length sample. From the intervals, we estimated the age of the variants and the credibility interval. The weighted average of the intervals where the weight is relative to the interval length was calculated. By doing so we assumed that the age is equally likely to lie anywhere within each interval and we gave equal weight to each of the 10,000 sampled branch lengths. The variant estimate was estimated to be the median of the probability density and the 95% credible interval was calculated as 2.5% and 97.5% quantiles. The age in generations was converted to years with the assumed generation time of 28 years per generation.