Blind and ad-hoc analysis of replicated experiments with the same conclusion for all experiments using GraphPad Prism v.9.5.0
All experiments were duplicated at least twice in a single experiment with the same conclusion. Each figure legend shows the exact number of replicated experiments. Samples and animals were randomly allocated to experimental groups and proceeded in experiments, and data were collected and analysed blind and ad hoc registered to groups or genotypes.
GraphPad Prism v.9.5.0 was used for statistical analysis. Mean and s.e.m. were used to represent a sample. Chi-squared and paired two-tailed Student’s t-tests were used to compare two samples; analysis of variance was used to compare multiple data points. No statistical methods were used to predetermine sample sizes.
Staining and clearing fat pad pads using a modified version of iDISCO for WAT, BAT and NPY+ cell counting
Whole fat pads were stained and cleared using a modified version of iDISCO as previously described18,46. In brief, WAT or BAT was dissected from mice perfused with 20 ml of PBS, and the tissues were fixed with 4% PFA overnight. Tissues were pretreated once each with 20, 40, 60 and 80% ethanol and twice in 100% methanol for 30 min, all at room temperature, and then bleached with 5% H2O2 diluted in 100% methanol for 24 h at 4 °C with shaking. After 30 min, the samples were rehydrated using 80, 60, 40 and 20%Methanol along with 10 g of heparin and 0.05% sodium azide in PBS. Then, the samples were put in a bath of water and blocked with a blocking buffer and permeabilizing solution (0% DMSO, 2% Triton X-100 and 3% PBS) at 37 C. For 5 days, the samples were put in a container with a 5% goat vaccine and a small amount of a drug called heparin in PBS. The tissues were put in 1% agarose, dehydrated in 20%, 40%, 60, 80 and 100% of methanol for an hour, and then put in a bottle of liquid for the night. After they sank and were then incubated in dibenzyl ether, they were free to explore. After the samples had been stored in dibenzyl ether, they were transferred toECi for x-rays. Images of cleared tissues were obtained with the use of a microscope; step size was 8 m, thickness was 3.98 m, horizontal dynamic focusing was set to eight steps, and exposure time was 180 ms. The samples were scanned under a 2/0.5 NA objective with a voxel size of 4.03/y and 8 m. The 488-mm laser with a 525-50 filter was the one used for AF488, while the other Lasers had a 620/60 filter and a 668-mm laser had a 680/30 filter.
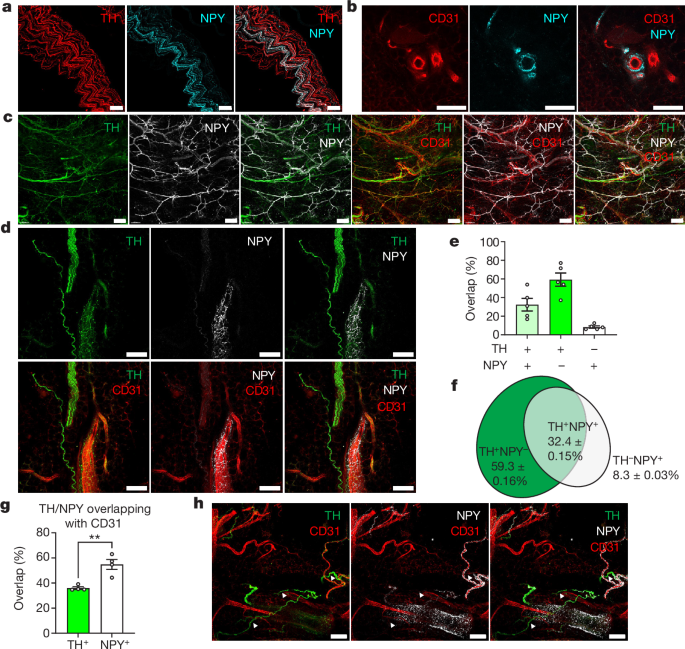
The percentages of the two types of cells were counted manually. A percentage of NPY+ neurons were counted manually in the hypothalamus. To calculate ganglionic mural cell coverage, 531, 531 and 30 μm xyz views were randomly picked and maximally projected to z using Fiji script. The areas were categorized by threshold and the coverage was calculated with area NPY+/ area CD31+.
The EdU+ cells percentage was counted with an unbiased method. Threshold was set automatically using the Otsu thresholding method.
Innervation of NPY+ axons was quantified using the ‘Surface’ tool in Imaris v.9.2. Labelled areas of NPY+ axons and CD31+ vessels in whole cleared iWAT were automatically segmented, and the innervation of NPY in vasculature was calculated as volumeNPY+/volumeCD31+. The coverage of DES+ mural cells in iWAT was calculated similarly, as volumeDES+/volumeCD31+. Confocal images showing NPY+ innervation were quantified using Fiji with an automatic unbiased method. The Otsu thresholding method was used to split the areaNPY+ and areaCD31+.
Overlapping between TH, NPY and CD31 was calculated using JACoP54, a Fiji plug-in47. To make this calculation, 472.33, 472.33 and 30 μm xyz regions were randomly picked and maximally projected to z using Fiji script. Labelled areas were automatically segmented using a threshold set by default, and overlapping percentages were calculated automatically by JACoP.
Source: Sympathetic neuropeptide Y protects from obesity by sustaining thermogenic fat
Structural cell isolation and culture using scRNA-seq datasets using Seurat v.4.2.0 and an array method to determine the size of mural cells
The public scRNA-seq datasets were analysed using the following method. The cells with fewer than 200 unique genes were discarded. After filtering, the gene x cell matrix was normalized using ‘NormalizeData()’ in Seurat v.4.2.0 (ref. The R contains 50 in it’s version. Data were then scaled using ‘ScaleData()’, and linear dimensional reduction performed by principal component analysis and calculation of UMAP coordinates for all cells using Seurat v.4.2.0. Cells were clustered using ‘FindNeighbour()’, with dimensions set to 15, and ‘FindClusters()’, with resolution set to 0.5. Each cluster was identified based on differentially expressed genes and known markers in the published literature5,22,29,51,52,53.
Mural cells were isolated and cultured following a modified version of the protocol previously published49. Mice were euthanized with a pen tobarbital and perfused with PBS to remove blood. The method used for isolating iBAT was described above. Cells were then seeded in 10 cm dishes with 10 ml of DMEM with 10% FBS for 2 days for the cells to attach to the bottom. Afterwards, 5 ml of the medium was replaced with mural cell-specific medium (DMEM with 2% FBS, supplied with 1% mural cell growth supplement (Science Cell, catalogue no. 1252)). Every other day, 5 ml of medium in dishes was refreshed until the cells grew confluent. For acquisition of pure mural cells, cells were labelled with an APC anti-CD104b antibody (1:100, BioLegend, catalogue no. 136007) and mural cells were sorted using the MojoSort magnetic sorting protocol with MojoSort buffer (BioLegend, catalogue no. 480017) and anti-APC nanobeads (BioLegend, catalogue no. 480071).
There were different kinds of cells differentiated to adipocytes. SVF and mural cells were isolated and seeded in 12-well plates at a density of 1 × 105 per millilitre. When the cells reached 95 % confluency, the cells were in beige cell medium with 10% family and 125 M dexamethasone. Maintenance medium was refreshed every 2 days. Total differentiation time for each well was 8 days. The NPY treatments in the experimental groups began when the medium was added. Oxygen consumption was measured using a Seahorse XF Analyzer.
The gift from R. Klemm to the University of Oxford was the 3T3-L1 preadipocyte cell line. The complete medium has high-glucose DMEM with glutamate and 10% calf serum. For subculture of preadipocytes the medium was transferred, and cells were washed with 1 ml of prewarmed 0.05% trypsin (ThermoFisher, catalogue no. 25300054) and digested with 200 μl of trypsin at 37 °C for 5–10 min. Afterwards, digestion was stopped and cells resuspended using the complete medium.
The cells were seeds at a higher rate than indicated by the concentrations ofPDGF-BB. 220-BB) and NPY (Cayman Chemical, catalogue no. CAY15071) were added to cells 24 h following cell seeding. The cells were collected within 5 days after the initial collection, fixed with 4% PFC or Trizol.
The following antibodies were used for fluorescent activated cell sorting (FACS) and flow cytometry: AF700 anti-CD45 (BioLegend, catalogue no. 103128), BUV395 anti-CD45 (BD Horizon, catalogue no. 564279), Pacific Blue anti-CD31 (BioLegend, catalogue no. 102421), APC anti-PDGFRa (BioLegend, catalogue no. 135907), AF488 anti-NG2 (Sigma, catalogue no. MAB5384A4) and AF488 anti-DES (abcam, catalogue no. ab185033, Y66). All the antibody for FACS were killed off at 1:500. The Live Fixable is near-IR. They were dead. The kit contains a cell stain and can be used to identify cells. Live staining was done for L10119).
In order to collect terminal blood, mice were euthanized and blood was collected from the left ventricle with a 25G needle and a 100 mM EDTA syringe. Blood was placed in tubes with 100 mM. After 15 min, the supernatant was put in fresh tubes with 50 M and aprotinin and they were able to separate plasma from blood cells. The concentration of NPY in mouse iWAT and blood plasma was determined using an NPY ELISA kit (Merck, catalogue no. EZRMNPY-27K). A FLUOstar Omega microplate reader was used to record the data.
The previously described proteins were taken from iWAT. In brief, dissected iWAT was placed in homogenizing tubes filled with 300 μl 50 mg−1 RIPA lysis buffer (Sigma, catalogue no. 20188) containing 50 μM DPP-IV inhibitor (Sigma, catalogue no. DPP4-010) and 500,000 IU ml−1 aprotinin (Sigma, catalogue no. A6103-1MG) and homogenized using a Precellys 24 homogenizer. Lipid in the tissue homogenize was removed by centrifuging at 20,000× rcf at 4 °C for 15 min, and the clear portion retained. The process was repeated three times to ensure complete removal of lipid.
Source: Sympathetic neuropeptide Y protects from obesity by sustaining thermogenic fat
Comparison of Thermogenic and Adipogenic Antigene Expression in CFX96 Real-Time PCR Detection System with TB Green Premix Ex Taq II
The data is from the CFX96 Real-Time PCR Detection System with the TB Green Premix Ex Taq II.
The Ct method uses a formula to calculate the relative expression of the Cttarget and Ctreference genes. The Ct method was used to compare the thermogenic and adipogenic genes. There are 5b and extended data figs. 5b,d,g, 9l,m and 10m.
The rat anti-CD31 and rat anti-PLVAP were used for staining. rabbit anti-NPY (Cell Signaling, catalogue no. D7Y5A, 1:500 dilution). ab30914, 1:500 dilution), chicken anti-TH (Aves Labs, catalogue no. TYH73787982, 1:500 dilution), rabbit anti-TH (Sigma, catalogue no. Ab152, 1:500 dilution), mouse anti-NPY1R (Santa Cruz, catalogue no. sc-393192, 1:200 dilution), rat anti-PDGFRα (BioLegend, catalogue no. 135902, APA5, 1:200 dilution), rabbit anti-TAGLN (abcam, catalogue no. ab14106, 1:250 dilution), Cy3 anti-αSMA (Sigma, catalogue no. C6198, 1A4, 1:250 dilution), goat anti-SOX17 (R&D, catalogue no. R&D catalogue no. of the anti-EPHB4 goat. AF3034, 1:250 dilution) and rabbit anti-UCP1 (abcam, catalogue no. ab10983, 1:500 dilution).
Source: Sympathetic neuropeptide Y protects from obesity by sustaining thermogenic fat
Simultaneous microscopy of epididymal white adipose tissue using lasers and two-photon excitation fluorescence
Animals were anaesthetized with ketamine (100 mg kg−1) and xylazine (10 mg kg−1) and a small incision was made to expose epididymal white adipose tissue for microscopy analysis. Intravital images of space resolution 1,024 × 1,024 pixels from epididymal WAT were obtained simultaneously using an optical microscope with coherent anti-Stokes Raman scattering, two-photon excitation fluorescence and a bright-field microscope, with a confocal LSM 780-NLO Zeiss inverted microscope Axio Observer Z.1 (Carl Zeiss). The procedures were performed at the National Institute of Science and Technology and the State University of Campinas. Two lines of lasers in wavelength pump and stokes were used to acquire adipocytes images and a second line of lasers in wavelength flex and stokes was used to acquire fluorescence images. T1164) In wavelength Stokes was 1,040. The video was started after the identification of blood vessels, and after ten frames, with 50 l (30 g) of diethylstilbestek (Dex tran) in it. T1162) was injected into the orbital plexus of the mouse. The video was recording for over 12 minutes. The intensity of fronescence was analysed for the whole video. The static image was analysed. For evaluation of tissue leakage, a region of interest was selected inside the vessel and another in the tissue (outside the vessel). The intensity ratio between the tissue and the vessel was calculated based on the florescence intensity in both areas. The blood flow was calculated using the average of the raw intensity of dextran within the artery over 100 s after stabilization of the signal.
Images were acquired using a Zeiss LSM880 confocal microscope with Zen-black software (v.2.1). A 10/0.45 NA objective was used to image samples, followed by a 20/0.8 NA objective. Solid-state lasers of 405, 561 and 633 nm and an Argon 488 laser were used for DAPI, AF488, AF546 and AF647 fluorophores, respectively.
For staining, whole-mount, PFA-fixed ganglia were dehydrated twice in 90, 70, and 30%ethanol and then rehydrated with 20 minutes of shaking at each concentration. Ganglia were then eaten with a 1.5 U level of dispase-1 and 300 g hyaluronidase. Ethanol-treated ganglia were blocked in blocking solution (3% bovine serum albumin, 2% goat serum, 0.1% Triton X-100 and 0.1% sodium azide in PBS) for 2 h and then incubated with primary antibodies diluted in the permeabilizing buffer for 3 days at 4 °C. ganglia were cultured for 3 more days, at 4 C, in the permeabilizing buffer. Finally, ganglia were dehydrated once each in 30, 50, 70 and 90% ethanol and twice in 100% ethanol then cleared using ethyl cinnamate (ECi). The cleared ganglia were mounted in a coverslip slide sandwich.
For example, if your sample is fixed it will be processed in 70%, 80%, and 90% of Ethanol and the other 75% xylene, and then in 60 C paraffin wax for each condition. Samples were embedded in paraffin wax, sectioned at 7 µm, mounted on coverslips and dried in an oven at 45–50 °C overnight. For 5 minutes, the samples were deparaffinized in xylene and then they had to be rehydrated in 95 and 80% ethanol and water. Adding 0.10% trypsin to each slide and incubation at 37 C for 20 minutes made the antigen retrieval possible. After washing with running water, samples were stained using the same procedure as for fixed cells.
For cryosection, samples were first embedded in OCT (VWR, catalogue no. 361603E) and frozen at −20 °C until cryosectioning. Samples were cut into sections and mounted on charged SuperFrost Plus slides. The same procedure is used for staining fixed cells.
The tissues were fixed in 4% para formaldehyde for at least four hours after they were separated from the mice’s tissues. Cells collected following in vitro experiments were washed once with PBS and fixed using 4% PFA for at least 4 h. The first stained avon bundle was first stained by incubation overnight with primary and secondary antibodies dissolved in the permeabilizing buffer. Cells were first treated with a permeabilizing buffer for 1 h at a room temperature and then stained at 4 C with primary anti-oxidants in the permeabilizing buffer. After being washed and stained with DAPI, the samples were put in a bin for 1 hour at room temperature. Sympathetic fibres or fixed cells were then mounted on slides with Fluoromount-G mounting medium (ThermoFisher, catalogue no. 00-4958-02).
The mice were fixed on a stereotaxic holder after being anaesthetized with avertin. Either AAV2/9-hSyn-Cre-EGFP-WPRE-pA (Taitool, catalogue no. Two 1,012 viral genomes, either S0230-9 or AAV2 9hSyn-EGFP-WPRE-pA. The nucleus tractus solitarius was injected with Taitool, two 1,012 virus genomes, 200 nl per side.
Source: Sympathetic neuropeptide Y protects from obesity by sustaining thermogenic fat
Long-term acclimatization of ThCre and Cx3cr1GFP/+ mice in a single housed Optris thermocamera
Animals between the ages of 12 and 14 weeks were used for thermoimaging. The data shown in Fig. 5e were recorded using an Optris thermocamera (Optris PI 160 with a standard 61 lens), and the data in Extended Data Fig. 10l were recorded using a FOTRIC 225 s infrared camera and analysed with FOTRIC software (v.5.0.8.214). Animals were single housed and placed under the thermocameras, with their back shaved to expose the skin above iBAT. Animals were acclimatized for four days with ad libitum access to food and water in individual cages under a 12-12 h light/dark cycle before a 14-h fast. The mouse had access to water during the fast.
iBAT temperature was recorded every 1 s over a 6-day period by storing the temperature of the warmest pixel in view using the software provided by the camera manufacturer (rel. 2.0.6, Optris PIX Connect). The average of IBAT temperatures was taken as a 1-h period.
The animals were analysed for their locomotor activity when they were 8 weeks old. Animals were kept in cages under controlled room temperature and humidity after a 12/12 h light/dark cycle with water and food. Data were collected for 72 h following a 1 day acclimatation period. The activity was recorded by using a software named Panlab. Animals aged 7–9 weeks were analysed for body composition using a Minispec LF50 (Brucker).
ThCre mice (B6.Cg-7630403G23RikTg(Th-cre)1Tmd/J, stock no. 008601) and Cx3cr1GFP/+ mice (Cx3cr1tm1Litt/LittJ, stock no. 008451) were purchased from the Jackson Lab. The I. Kalajzic Laboratory donated the mice under the material transfer agreement. The tissues of NPY-GFP mice were procured from the Brandy Memorial Laboratory at Yale University. The Department of otology-Head and Neck Surgery at the University of Michigan received Tissues from Npy1rCre;Rosa26tTomatod mice. The NPY-cKO mice were created by crossing the ThCre and Npy Flox mice. Diet-induced obesity was achieved by feeding mice an HFD (Diet Research, product no. D12492) when they were 7 weeks old, and this feeding regime lasted for 10 weeks. The body weight of each mouse and food consumption in each cage were recorded weekly. All mice were group housed in standard housing under controlled room temperature (21–23 °C) and 50% humidity under a 12/12 h light/dark cycle and given access to diet and water ad libitum. The project licence provided for all experimental procedures to be conducted on living animals. Personal licences were granted by the UK Home Office. The University of Oxford has an Ethical Review Panel.
Using the AneuFinder cDNA Detector to Identify Single-embryo Cell Divisions Producing De Novo Chromosome Aberrations
Experimental reproducibility was demonstrated as follows: Fig. 1g, two independent experiments; Fig. 2d, two (2-cell) and three (1-cell, 4-cell) independent experiments; Fig. Two independent experiments are described in the figure. 3b, two independent experiments; Fig. 3c, four independent experiments; 3g, two 2 to-4-cell and 16- to-32-cell experiments; and 3h, two independent experiments. 3i, two independent experiments; Fig. 4a–d, two independent experiments; Fig. 4h, two independent experiments; Fig. 4i, two independent experiments; and Fig. 4j, four independent experiments.
The findCNGs command in AneuFinder has a 500 kb bin Size, so we used it to analyse the chromosomes. Using scRepli-seq data of 4-, 8- and 16-cell-stage embryos, we determined the cell division that produced de novo chromosome errors as follows. The embryo had two cells that had a 3:1 copy-number ratio, but they were judged to be a pair of sister cells that had a chromosome segregation error. 3b as de novo chromosome aberrations). Errors in the divisions preceding the last division were seen as evidence that the cells had experienced identical chromosomal abnormality. Details of all of the detected chromosome aberrations are shown in Extended Data Fig. 6b and Supplementary Table 1. Since we pooled embryo samples from multiple females before single-embryo sampling, our dataset is not compatible with female-to-female variability assessment.
Principal component 1 (PC1; A/B compartment profile) of Hi-C data in 200 kb bins was computed from the .hic file using published mapped Hi-C datasets of sperm68 and cumulus60 cells as described previously16. The genomic coordinates of PC1 profiles were converted from mm10 to mm9 using the liftover tool (UCSC Genome Browser). The four PC1 categories we defined are A1, A2, B2 and B1 and contained 25% of all PC1 values.
After linearization of the template plasmid, mRNA was synthesized using the mMESSAGE mMACHINE KIT (Ambion). Synthesized RNAs were stored at −80 °C until use. The stem cells were in a ratio of l1 to l1 Major-satellite. Live-cell imaging was performed as previously described66 with some modifications. The Carl Zeiss 40 C-Apochromat 1.2NA water-immersion objective lens can be used to control a confocal microscope. At 2min 15s intervals, 17 confocal sections (each 1.5 m) of 512 xy images were acquired for major-satellite imagery. Each 2 m section of the 512 512 Xy images was acquired at 3 min intervals for a total of at least 10 h. There were 29 confocal Z sections for PCNA with a total volume of 84.85 84.80 83.50 m. In Figs. 3c,g and 4j and Extended Data Fig. To image blastomeres that were just entering M phase and close to the objective lens, we chose them and chose between two and three blasts per embryo.
For the experiments described in Extended Data Fig. MC12 cells were treated with 0.25 M nocodazole for 17 h and then subjected to 3 liters of poison ivy treatment for 3 h. After the cells were cultured in the presence of 10 mM 2-AP, they were cultured again in 15 h with or without the drug. After removing aphidicolin or 2-AP, the cells were labelled with 20 μM EdU for 60 min and stained with EdU. The C57BL/6 strain 4-cell and 8-cell embryos within 1–2 h after the 2-to-4-cell or 4-to-8-cell division (that is, in early S phase) were treated with 3 μg ml−1 aphidicolin with or without 2-AP for 5.5 h followed by reagent removal and EdU staining.
Source: Embryonic genome instability upon DNA replication timing program emergence
Fork speed measurement using Alexa Fluor 647 ssDNA signals and a method to determine the mobile class forks in a DNA chip
The image of a DNA chip was subjected to auto-thresholding in the software. Individual DNA fibres were identified as a series of linearly arranged Alexa Fluor 647 (ssDNA) ‘dot’ signals (Fig. 2h). The thresholded fibres containing both Alexa Fluor 488 (IdU+CldU) and Alexa Fluor 594 (CldU) signals were manually categorized into those with ‘immobile’, ‘intermediate’ and ‘mobile’ class forks. The immobile class forks were defined as those with only one dot signal from IdU to CldU, and no gaps between it. Here, gaps were defined as those with at least one dot of Alexa Fluor 647 ssDNA signal (Fig. 2h (immobile)). The mobile class forks were defined as those with a series of dot signals (≥2 dots) of IdU + CldU The dot signals have a series on either side of the same DNA fibre. The intermediate class forks were defined as those with an intermediate character between the two other classes; these fibres also contained dot signals of IdU + CldU and CldU with gaps between dots but also contained some IdU + CldU single colour signals (that is, IdU-only regions) in between these dot signals. There is a method for measuring the mobile fork class fibres in the IOD method. 5b. The IOD between the immobile forks was calculated using the distance between the dots that is the most bright in the centre of the dots. To determine the fork speed of the mobile forks, CldU tracks flanked by IdU tracks were identified, their lengths were measured, and were divided by the duration of the second pulse (30 min). CldU tracks that had no ssDNA signals ahead of them were excluded from the fork speed measurement as these fibres may have been broken in the middle of the CldU track. There is a Supplementary Note 2 for the immobile fork speed measurement.
We obtained the signal intensity for H3K9me2, H2AX and p-CHK1 within the nucleus to measure the levels of histone H3. We then subtracted the mean cytoplasmic signal intensity (Ime_cyto), which was obtained from a region near the nuclei, from the Ime_nuc value (Ime_nuc − Ime_cyto). We determined the H3 level in the same way. Finally, we calculated the ratio between the two values (Ime_nuc − Ime_cyto)/(IH3_nuc − IH3_cyto).
The embryo were fixed with 2% paraformaldehyde, which is in PBS-polyvinyl alcohol. After blocking and permeabilization in PBS-PVA, the embryos were washed multiple times and incubated with appropriate primary antibodies. The sample was taken with 40 g of Hoechst 33342. Finally, the embryos were washed and transferred to BSA-PVA for imaging with a Zeiss LSM780 confocal microscope. The following primary antibodies were used: mouse anti-γH2A.X (phosphorylated Ser139) (1:200, Abcam, ab22551); rabbit anti-histone H3 (1:200, Abcam, ab62706); mouse anti-histone H3 (1:200, Abcam, ab195277); mouse anti-histone H3K9me2 (1:200, Monoclonal Antibody Institute, Japan (MABI), MABI0317); rabbit anti-phosphorylated-CHK1 (Ser345) (133D3) (1:200, Cell Signaling Technology, 2348S) antibodies. The secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG (H+L) (A11029); goat anti-rabbit IgG (H+L) (A11034); Alexa Fluor 555 goat anti-mouse IgG (H+L) (A21424) (1:400, Invitrogen).
scRepli-seq experiments using mouse embryos were performed as previously reported with slight modifications. In brief, unfixed single blastomeres (derived from BDF1 or B6MSM strain embryos) were collected into 8-well PCR tubes with 6 μl cell lysis buffer (288 μl of H2O, 2 μl of 10 mg ml−1 proteinase K (Sigma-Aldrich, P4850), 32 μl of 10× single-cell lysis and fragmentation buffer (Sigma-Aldrich, L1043)), incubated at 55 °C for 1 h and then at 99 °C for 4 min for gDNA isolation and fragmentation. We analysed all blastomeres of the embryo only if there was damage to the sample. For scRepli-seq experiments after EdU staining (Extended Data Fig. After taking the photographs of EdU-stained cells, the cells were collected by mouth pipette and placed in 12l of cell lysis buffer and 55 C for 16 hours. Next-Generation Sequencing library construction kit, KAPA, was performed according to the instructions of the manufacturer. The samples were processed for NGS on the Illumina Hiseq 1500 or Hiseq X Ten system (80-bp-length single-read or 150-bp-length paired-end read sequencing).
SCNT was performed as previously described61. Groups of MI I oocytes were transferred into the droplets of M2 medium to be collected on the microscope stage. Oocytes were held with a pipette. A hole was made in the zona pellucida through the application of several piezo-pulses using an enucleation pipette. The MII spindle was aspirated into the pipette with a minimal volume of ooplasm, and the resulting enucleated oocytes were transferred into CZB. For nuclear injection, donor cumulus cells were gently aspirated in and out of the injection pipette to broken plasma membranes. The reconstructed oocytes were kept in the incubator while the nucleus was injected. Reconstructed oocytes were parthenogenetically stimulated by incubation in CZB supplemented with 10 mM SrCl2, 2 mM ethylene glycol tetraacetic acid (EGTA), and 5 µM latrunculin A for 10 h, and then cultured in CZB.
All animal experiments conformed to the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation of the RIKEN Center for Biosystems Dynamics Research. B6D2F1 (C57BL/6 × DBA/2) and C57BL/6 mice, aged 8–10 weeks, were used to produce oocytes and sperm. For allele-specific analysis, C57BL/6 female mice and MSM/Ms male mice were used to produce oocytes and sperm, respectively. To eliminate the effect of individual differences between mice, multiple mice were used in each experiment as follows. 2 mice, 4 mice, 8 mice, 16 mice, and 4 mice; Figure 1b. 1d, 10 mice each (1-cell, 2-cell and 4-cell); 2d, 4 (1-cell), 3 (2-cell) and 3 (4-cell) mice are shown. 3b, 4 in vitro and 2 in vivo (2-to-4-cell), 8 in vitro and 7 in vivo (4-to-8-cell), and 4 in vitro and 2 in vivo (8-to-16-cell) mice; Fig. 3c, 15 mice each (2-to-4-cell, 4-to-8-cell and 8-to-16-cell); Fig. 3g, 3 mice each (2-to-4-cell, 4-to-8-cell, 8-to-16-cell and 16-to-32-cell); Fig. 3h, 4 (2-cell) and 3 (4-cell and 8-cell) mice; Fig. 3i, 5 (2-cell, 4-cell, and 8-cell) mice; Fig. 4d, 4 mice; Fig. 4h–i, 6 mice; Fig. 4j, 16 mice; Extended Data Fig. 7a, 7 (2-to-4-cell DMSO), 6 (2-to-4-cell aphidicolin 15 ng ml−1, 30 ng ml−1, and 60 ng ml−1), 8 (2-to-4-cell aphidicolin 75 ng ml−1), 16 (4-to-8-cell DMSO), 6 (4-to-8-cell aphidicolin 15 ng ml−1, 30 ng ml−1 and 60 ng ml−1), 6 (4-to-8-cell aphidicolin 75 ng ml−1), 12 (8-to-16-cell DMSO), 8 (8-to-16-cell aphidicolin 15 ng ml−1, 30 ng ml−1 and 60 ng ml−1) and 6 (8-to-16-cell aphidicolin 75 ng ml−1) mice.